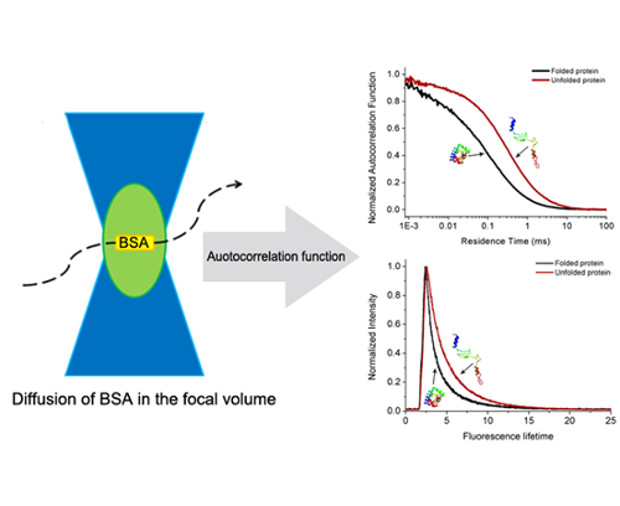
The structural dynamics of proteins is crucial to their biological functions. A precise and convenient method to determine the structural changes of a protein is still urgently needed. Herein, we employ fluorescence correlation spectroscopy (FCS) to track the structural transition of bovine serum albumin (BSA) in low concentrated cationic (cetyltrimethylammonium chloride, CTAC), anionic (sodium dodecyl sulfate, SDS), and nonionic (pentaethylene glycol monododecyl ether, C12E5 and octaethylene glycol monododecyl ether, C12E8) surfactant solutions. We find that the diffusion coefficient of BSA decreases abruptly with the surfactant concentration in ionic surfactant solutions at concentrations below the critical micelle concentration (CMC), while it is constant in nonionic surfactant solutions. According to the Stokes-Sutherland-Einstein equation, the hydrodynamic radius of BSA in ionic surfactant solutions amounts to ~6.5 nm, which is 1.7 times larger than in pure water or in nonionic surfactant solutions. The interaction between BSA and ionic surfactant monomers is believed to cause the structural transition of BSA. We confirm this proposal by observing a sudden shift of the fluorescence lifetime of ATTO-BSA in ionic surfactant solutions at the concentration below CMC. No change in the fluorescence lifetime is detected in nonionic surfactant solutions. Moreover, by using FCS we are also able to identify whether the structural change of protein results from its self-aggregation or unfolding. You can read more details in Soft Matter, 2015, 11, 2512.